CLEM-Reg
An automated point cloud based registration algorithm for correlative light and volume electron microscopy
this posts summarises the paper I helped work on during my placement year at the Francis Crick Institute.
Background
Correlative light and electron microscopy (CLEM) is a powerful imaging technique that combines the advantages of fluorescence microscopy (FM) and electron microscopy (EM). FM allows you to visualise fluorescently labeled proteins in living cells and tissues, while EM provides ultra-structural details at much higher resolutions. However, aligning the FM and EM image volumes is challenging due to their different scales, appearances, and potential deformations during sample preparation. Traditionally, expert microscopists have manually identified corresponding landmarks between the two imaging modalities and aligned the datasets through computationally intensive optimization methods. This manual process is extremely laborious, low-throughput, and can introduce human bias.
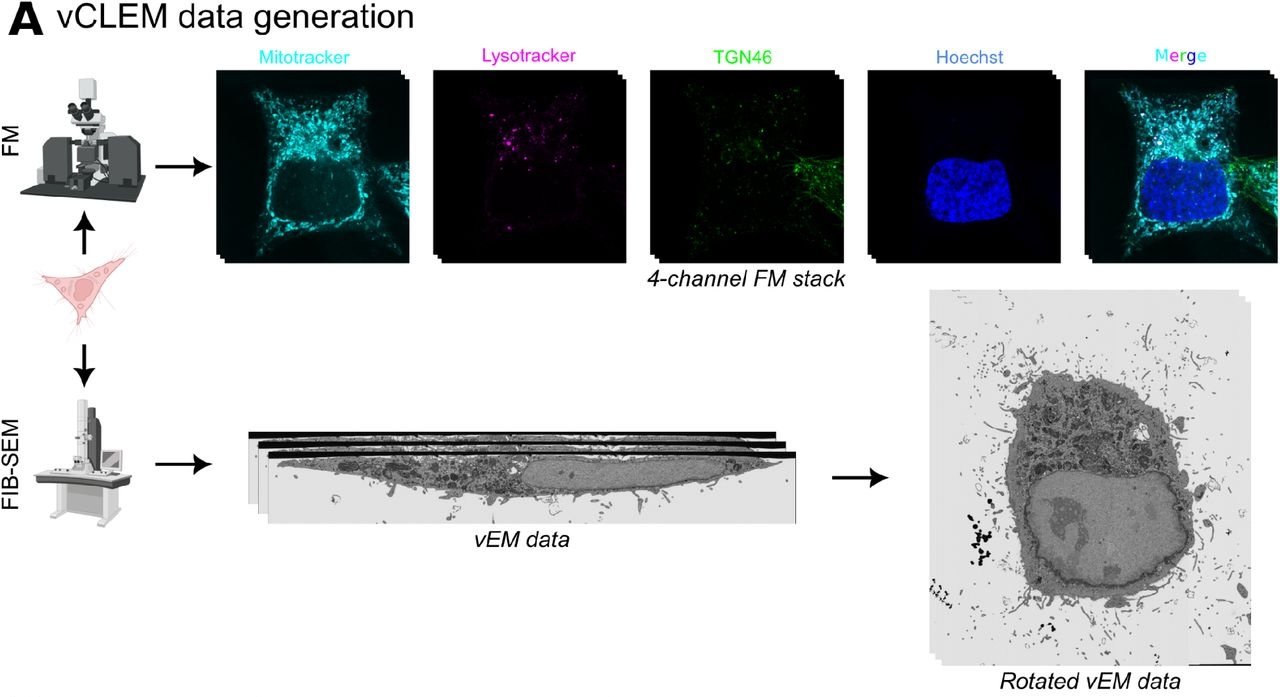
Methods
To address the limitations associated with current techniques for registration of CLEM image volumes CLEM-Reg was developed, a fully automated workflow to align CLEM datasets. The key innovation is using mitochondria as internal landmarks that can be automatically segmented in both FM and EM images using a combination of classical image processing and deep learning techniques. Once the mitochondria are segmented, CLEM-Reg represents them as sparse 3D point clouds - a modality-agnostic representation. These point clouds are then aligned using coherent point drift registration, a state-of-the-art probabilistic algorithm. The final alignment transformation is used to warp the FM volume onto the EM volume, producing the integrated CLEM overlay.
Results
Tested CLEM-Reg on three newly acquired benchmark CLEM datasets of HeLa cells revealed its performance is on par with
manual expert alignments. Remarkably, CLEM-Reg achieved near expert-level registration accuracy in aligning target structures like lysosomes and endosomes that were not used as landmarks. One of the major advantages of CLEM-Reg is that it entirely automates the alignment process in an unbiased and reproducible manner, eliminating the need for tedious manual input. It is also implemented as an easy-to-use plugin for napari, a popular open-source multi-dimensional image viewer.
Conclusions
Overall, CLEM-Reg represents a significant advance that could enable wider adoption of CLEM by increasing throughput and reproducibility. The paper’s authors have made their code and the benchmark datasets publicly available to aid further development of multimodal registration algorithms by the wider scientific community.